Brain Tumor Classification with Docker Compose and PyTorch Lightning
This project demonstrates how to set up training, evaluation, and inference for Brain Tumor Classification using Docker and PyTorch Lightning. We employ Docker containers for environment consistency, and PyTorch Lightning for structured, modular training, evaluation, and inference processes. The dataset used in this project is the Brain Tumor Classification (MRI) dataset, managed through Docker Compose.
Table of contents
Requirements
This project requires the following packages to be installed:
PyTorchtorchvisionPyTorch LightningNumPyscikit-learnTensorBoardMatplotlib
Dataset
The dataset used in this project is the Brain Tumor Classification (MRI). It consists of MRI scans categorized into four classes: 👇
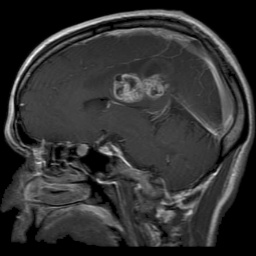 Glioma Tumor | 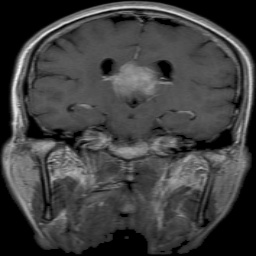 Meningioma Tumor |
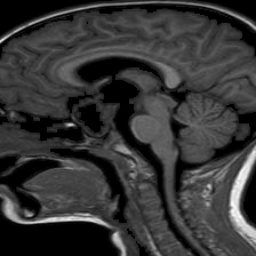 Normal | 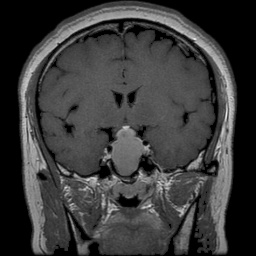 Pituitary Tumor |
Each class is stored in separate directories, allowing for easy management and access during the training and evaluation process.
Why PyTorch Lightning?
PyTorch Lightning simplifies the training process by modularizing the code and separating concerns. It helps you focus more on the logic of your models instead of boilerplate code.
✅ Simplified Code Structure: It encourages clean and organized code, making it easier to maintain and scale.
✅ Flexibility: It allows for an easy switch between different training strategies (such as multi-GPU training, TPU support, etc.) with minimal changes.
✅ Built-in Features: It includes built-in logging, checkpointing, and early stopping, reducing manual implementations.
Docker Setup
DevContainer
The .devcontainer setup allows you to develop in a pre-configured environment using VS Code:
{
"name": "Brain Tumor Classification",
"dockerFile": "Dockerfile",
"settings": {
"terminal.integrated.shell.linux": "/bin/bash"
},
"extensions": [
"ms-python.python",
"ms-azuretools.vscode-docker"
],
"postCreateCommand": "pip install -r requirements.txt"
}
Docker Compose Services
The Docker Compose file defines three services: train, evaluate, and infer:
version: '3.8'
services:
train:
# Train service
build:
context: .
dockerfile: Dockerfile.train
shm_size: "2gb"
volumes:
- host:/opt/mount
- ./model:/opt/mount/model
- ./data:/opt/mount/data
evaluate:
# Evaluate service
build:
context: .
dockerfile: Dockerfile.eval
volumes:
- host:/opt/mount
- ./model:/opt/mount/model
- ./data:/opt/mount/data
infer:
# Inference service
build:
context: .
dockerfile: Dockerfile.infer
volumes:
- host:/opt/mount
- ./model:/opt/mount/model
- ./data:/opt/mount/data
- ./predictions:/opt/mount/results
volumes:
host:
Training and Evaluation
To set up and execute the training, evaluation, and inference processes, follow these steps:
1️⃣ Build Docker Images:
-
Build the Docker images for all services using:
docker compose build
2️⃣ Train the Model:
-
To train the model, run:
docker compose run train -
The
BrainTumorClassifierwill train the model and save the checkpoints in the shared volume.
3️⃣ Evaluate the Model:
-
To evaluate the model, execute:
docker compose run evaluate -
This will load the saved checkpoint and print the validation metrics using
eval.py.
Inference
To run inference on sample images:
docker compose run infer
- The
infer.pyscript will predict the labels for 10 random images and save the output to thepredictionsdirectory.
Each service uses volume mounts to ensure that data, checkpoints, and results are accessible across different containers, maintaining consistency.
Results
Check the predictions in the predictions folder. Below are some sample results:
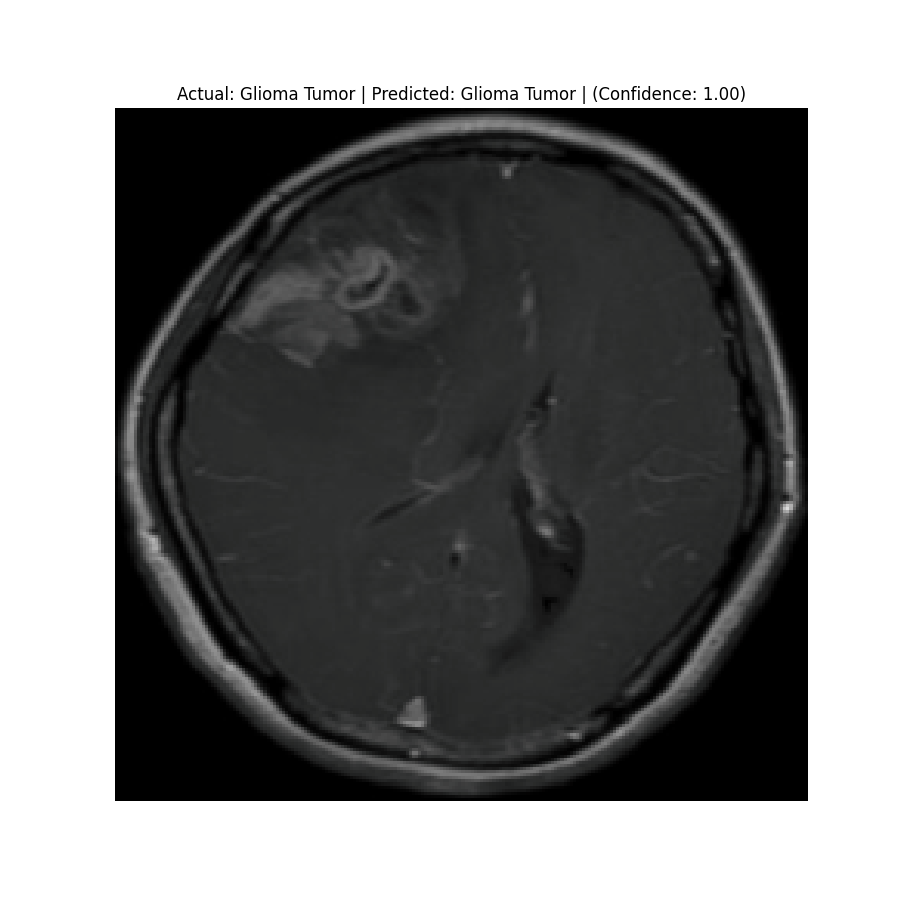 Actual: Glioma Tumor Predicted: Glioma Tumor (Confidence: 1.00) | 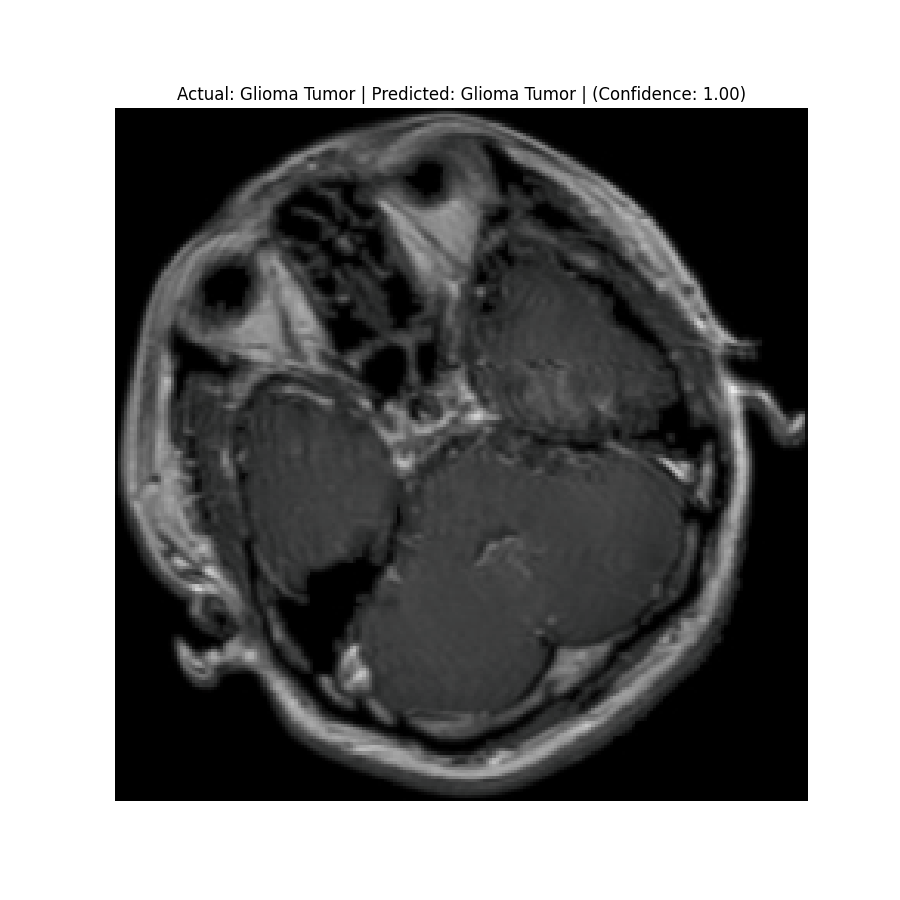 Actual: Glioma Tumor Predicted: Glioma Tumor (Confidence: 1.00) |
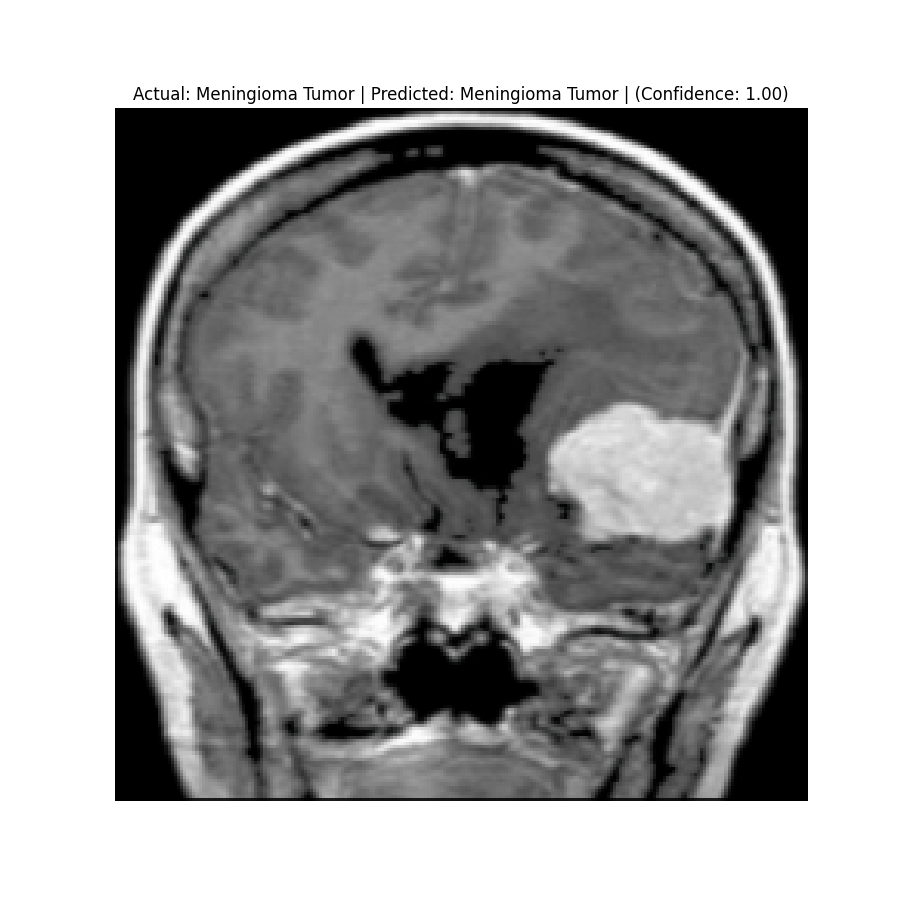 Actual: Meningioma Tumor Predicted: Meningioma Tumor (Confidence: 1.00) | 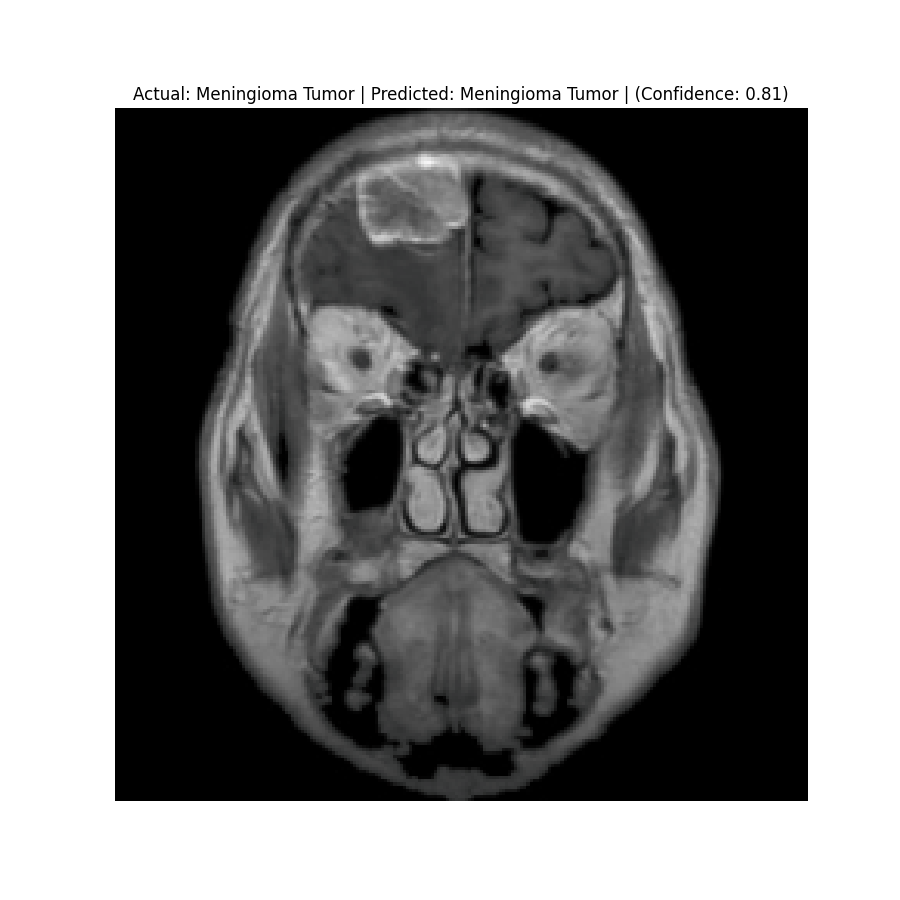 Actual: Meningioma Tumor Predicted: Meningioma Tumor (Confidence: 0.81) |
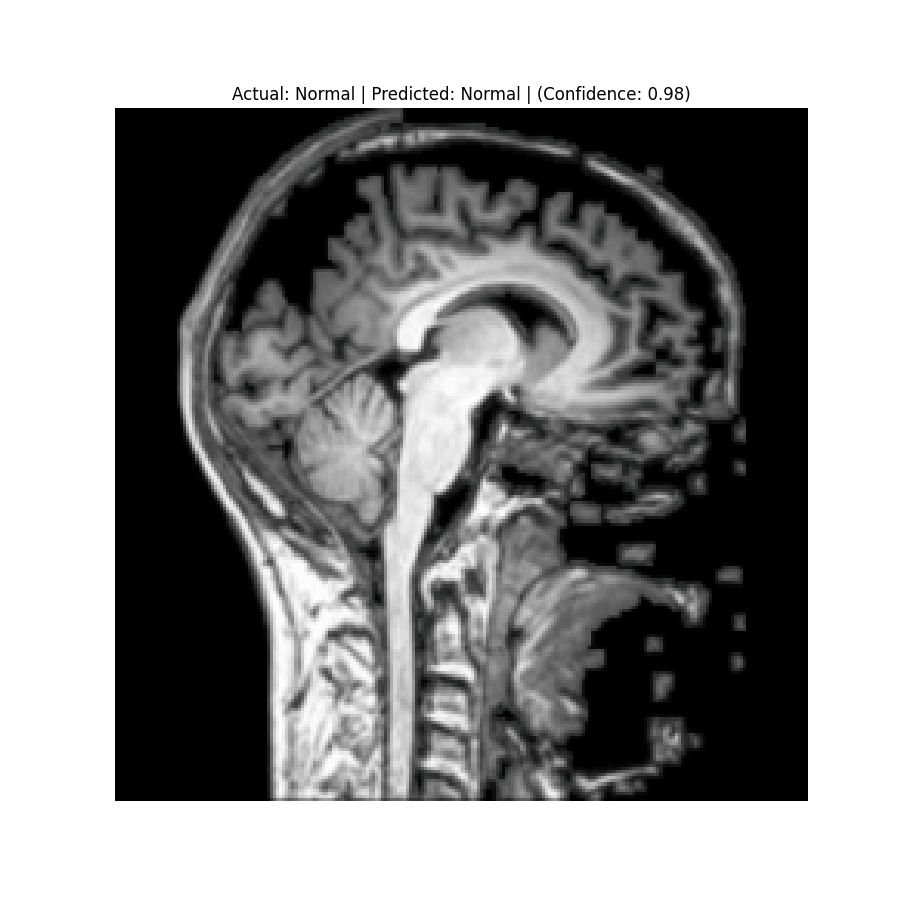 Actual: Normal Predicted: Normal (Confidence: 0.98) | 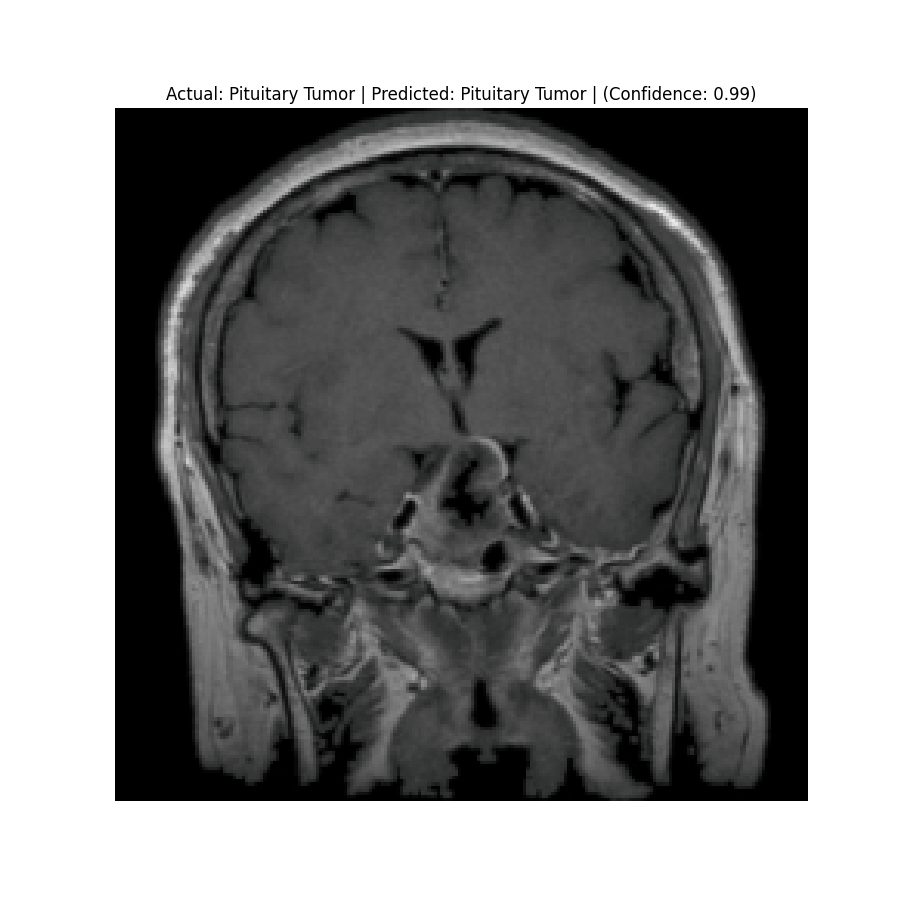 Actual: Pituitary Tumor Predicted: Pituitary Tumor (Confidence: 0.99) |
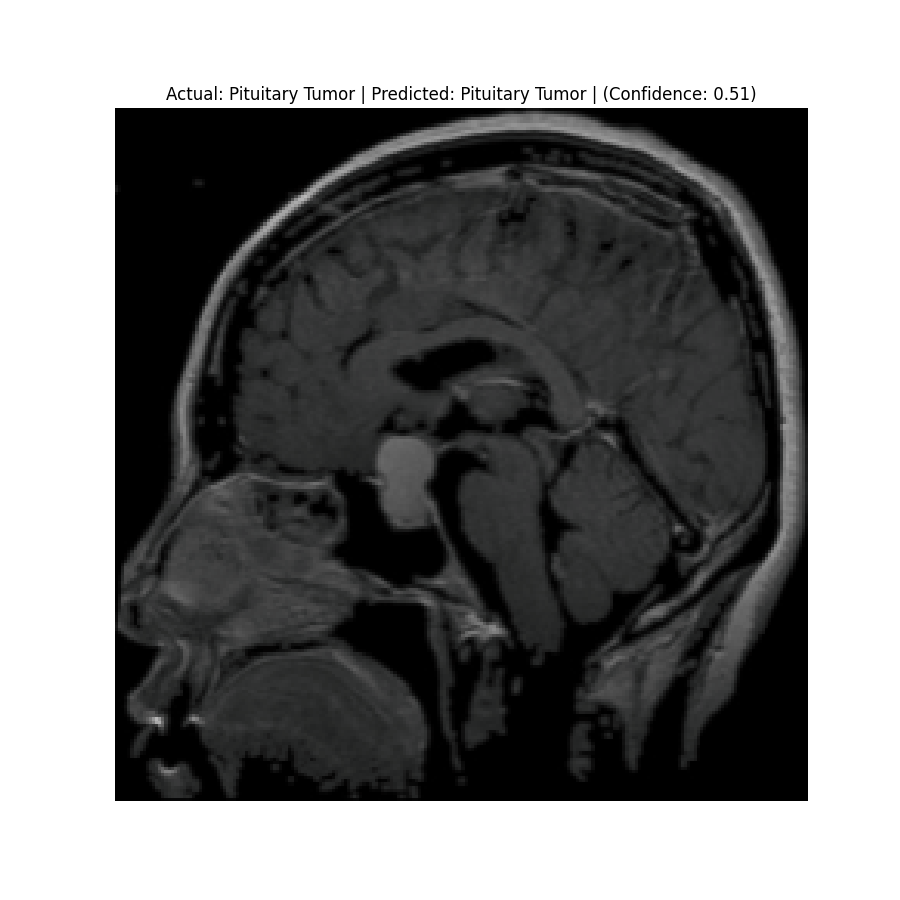 Actual: Pituitary Tumor Predicted: Pituitary Tumor (Confidence: 0.51) | 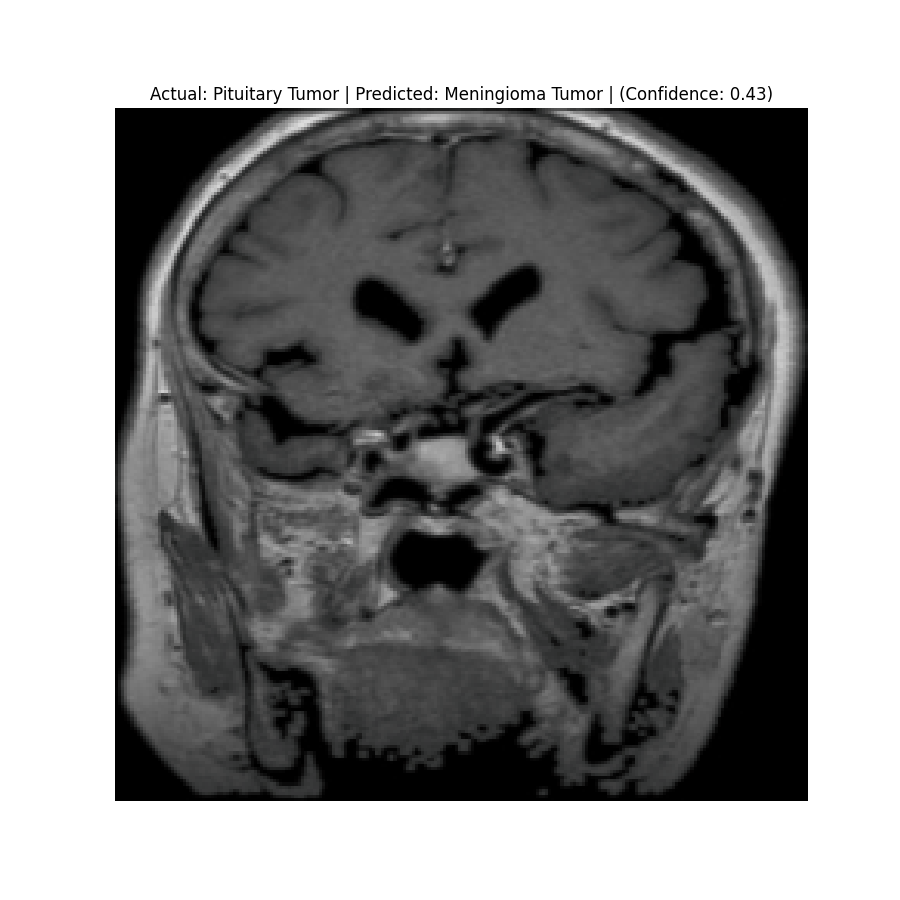 Actual: Pituitary Tumor Predicted: Meningioma Tumor (Confidence: 0.43) |